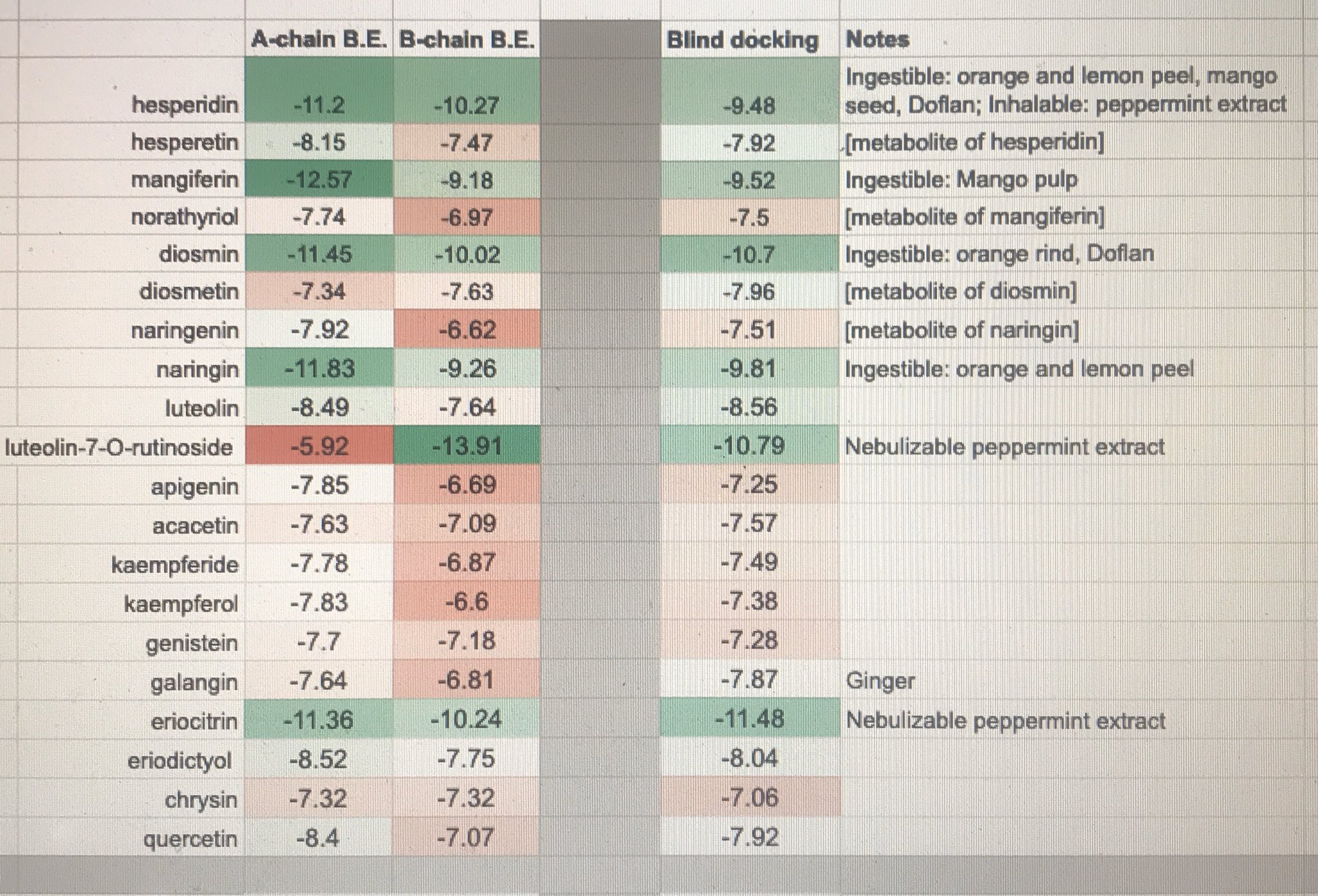
In Silico effect of Flavonoids on SARS-CoV 3CLpro progressively targeting A and B-chains' HIS41-CYS145 interfaces
Results:
Micro-discussion:
So while I've been a very strong proponent of Hesperidin based on in vitro results. benign cytox values, inherent known safety, and wide suppy availability, this Autodock Vina docking study shows stronger hits in 1. Hinokiflavone, 2. Luteolin-O-7-rutinoside, 3. Eriocitrin, 4. Sulcatone A, and 5. Amentoflavone.
As amentoflavone is the only one of the above with known outperforming in-vitro efficacy demonstration on SARS-CoV 3CLpro, while showing two orders of magnitude margin for cytotoxicity on Vero cells, I advocate that in vitro trials targeting phytotherapies focus on amentoflavone as applied to (e.g. e.coli-expressed) SARS-CoV-2 3CLpro proteins.
Source data:
https://docs.google.com/spreadsheets/u/1/d/19RGHp9ls3fdjDPYgT-YtNwWXsMwI9f3Glqz6KnedK0M/edit?usp=drive_web&ouid=117587353864850994495
PDB File: 2duc.pdb
Ligand Files: PubChem 3D Conformers
Citations:
Yu Wai Chen, Chin-Pang Bennu Yiu, Kwok-Yin Wong, (Hong Kong Polytechnic University)
“Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidate”, F1000Research, 2020
O. Trott, A. J. Olson,
“Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidate”, F1000Research, 2020
O. Trott, A. J. Olson,
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, Journal of Computational Chemistry 31 (2010) 455-461 DOI 10.1002/jcc.21334
“Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds", Antiviral Research, Volume 68, Issue 1, October 2005, Pages 36-42.
Ryu, YB et al. (Korea Research Institute of Bioscience and Biotechnology)
"Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition.", Bioorg Med Chem., 2010 Nov 15; 18(22):7940-7.
DOI 10.1016/j.bmc.2010.09.035
Michel F. Sanner.
Python: A Programming Language for Software Integration and Development. J. Mol. Graphics Mod., 1999, Vol 17, February. pp57-61
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S. and Olson, A. J. (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Computational Chemistry 2009, 16: 2785-91